Plot static litre plots.
plotLitre( data = data, dataMetrics = NULL, dataSE = NULL, geneList = NULL, threshVar = "FDR", threshVal = 0.05, option = c("hexagon", "allPoints"), pointSize = 2, pointColor = "orange", xbins = 10, outDir = tempdir(), saveFile = TRUE )
Arguments
| data | DATA FRAME | Read counts |
|---|---|
| dataMetrics | LIST | Differential expression metrics; default NULL |
| dataSE | SUMMARIZEDEXPERIMENT | Summarized experiment format that can be used in lieu of data and dataMetrics; default NULL |
| geneList | CHARACTER ARRAY | List of ID values of genes to be drawn from data as litre plots. Use this parameter if you have predetermined genes to be drawn. Otherwise, use dataMetrics, threshVar, and threshVal to create genes to be drawn; default NULL |
| threshVar | CHARACTER STRING | Name of column in dataMetrics object that is used to threshold significance; default "FDR" |
| threshVal | INTEGER | Maximum value to threshold significance from threshVar object; default 0.05 |
| option | CHARACTER STRING ["hexagon" | "allPoints"] | The background of plot; default "hexagon" |
| pointSize | INTEGER | Size of plotted points; default 2 |
| pointColor | CHARACTER STRING | Color of gene superimposed on litre plot; default "orange" |
| xbins | INTEGER | Number of bins partitioning the range of the plot; default 10 |
| outDir | CHARACTER STRING | Output directory to save all plots; default tempdir() |
| saveFile | BOOLEAN [TRUE | FALSE] | Save file to outDir; default TRUE |
Value
List of n elements of litre plots, where n is the number of genes determined to be superimposed through the dataMetrics or geneList parameter. If the saveFile parameter has a value of TRUE, then each of these litre plots is saved to the location specified in the outDir parameter as a JPG file.
Examples
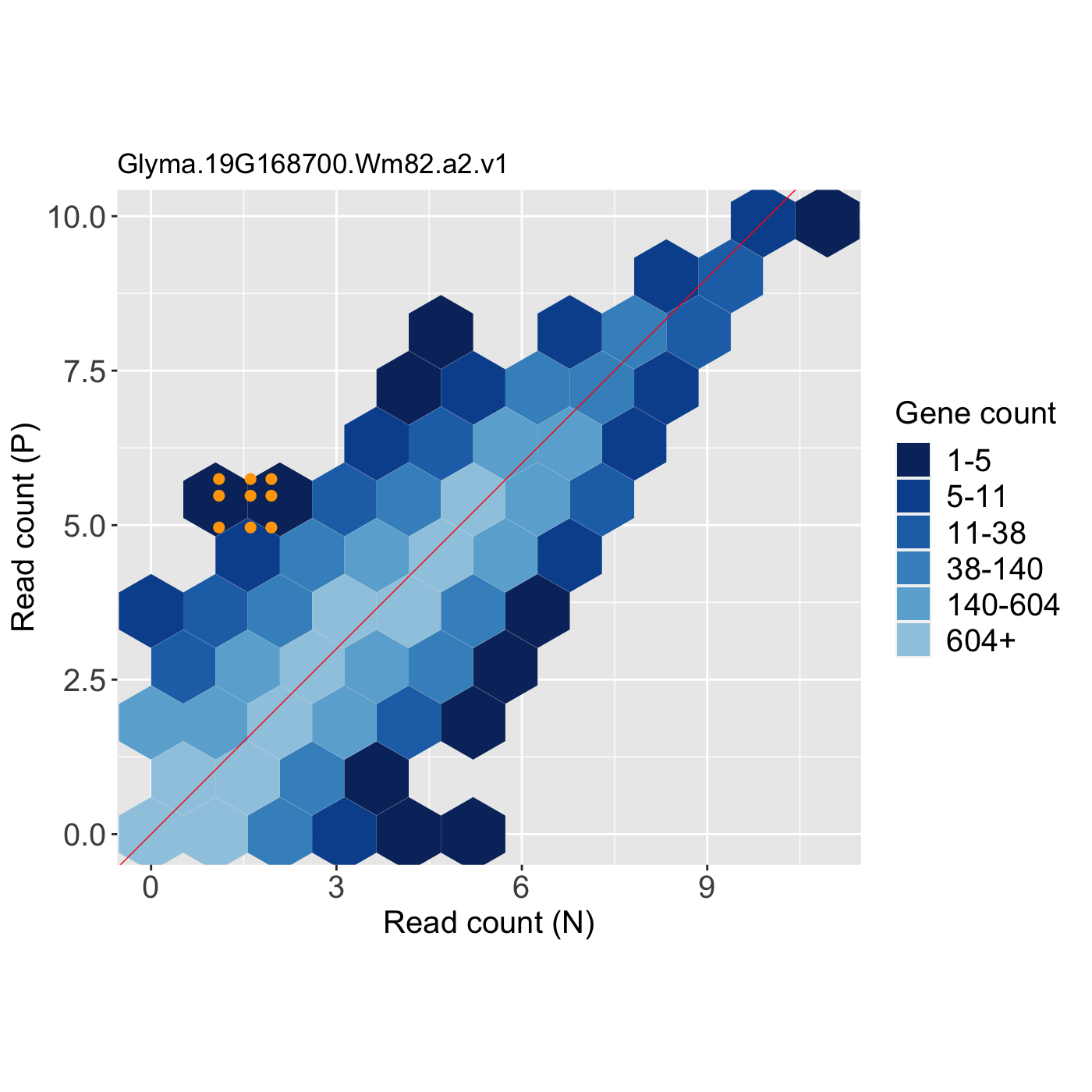
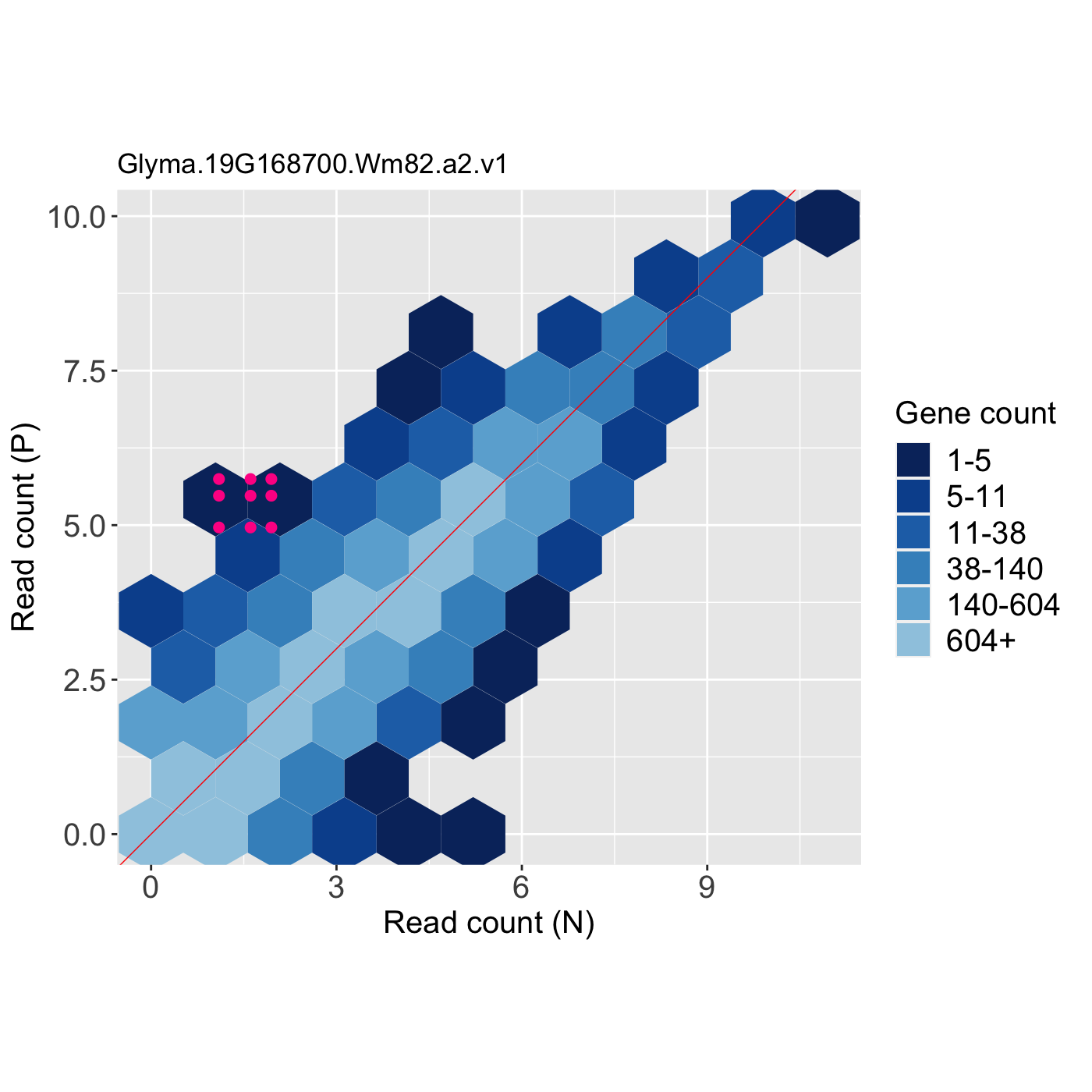
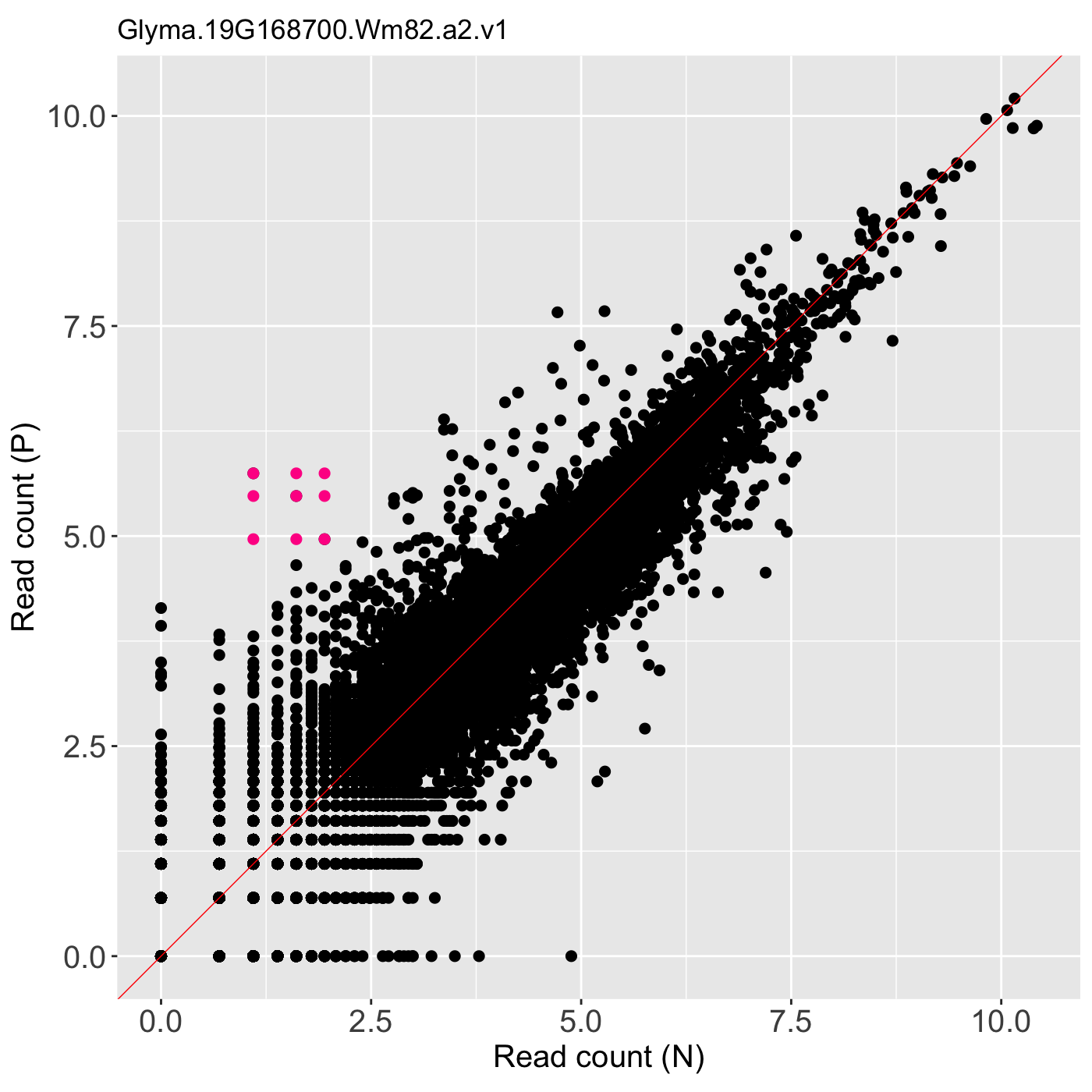
# The first set of three examples use data and dataMetrics # objects as input. The last set of three examples create the same plots now # using the SummarizedExperiment (i.e. dataSE) object input. # Example 1: Create litre plots for each of the 61 genes with FDR < 1e-10. # Examine the first plot (gene "N_P_Glyma.19G168700.Wm82.a2.v1") data(soybean_ir_sub) soybean_ir_sub[,-1] <- log(soybean_ir_sub[,-1]+1) data(soybean_ir_sub_metrics) ret <- plotLitre(data = soybean_ir_sub, dataMetrics = soybean_ir_sub_metrics, threshVal = 1e-10, saveFile = FALSE) length(ret)#> [1] 61#> [1] "N_P_Glyma.19G168700.Wm82.a2.v1"ret[[1]]# Example 2: Create litre plots for each of the five most significant genes # (low FDR values). View plot for gene "N_P_Glyma.19G168700.Wm82.a2.v1". geneList = soybean_ir_sub_metrics[["N_P"]][1:5,]$ID ret <- plotLitre(data = soybean_ir_sub, geneList = geneList, pointColor = "deeppink") names(ret)#> [1] "N_P_Glyma.19G168700.Wm82.a2.v1" "N_P_Glyma.13G293500.Wm82.a2.v1" #> [3] "N_P_Glyma.05G188700.Wm82.a2.v1" "N_P_Glyma.13G173100.Wm82.a2.v1" #> [5] "N_P_Glyma.11G141800.Wm82.a2.v1"ret[["N_P_Glyma.19G168700.Wm82.a2.v1"]]# Example 3: Create one litre plot for each of the five most significant # genes (low FDR values). View the plot for gene # "N_P_Glyma.19G168700.Wm82.a2.v1". Use points instead of the default # hexagons as the background. ret <- plotLitre(data = soybean_ir_sub, geneList = geneList, pointColor = "deeppink", option = "allPoints") names(ret)#> [1] "N_P_Glyma.19G168700.Wm82.a2.v1" "N_P_Glyma.13G293500.Wm82.a2.v1" #> [3] "N_P_Glyma.05G188700.Wm82.a2.v1" "N_P_Glyma.13G173100.Wm82.a2.v1" #> [5] "N_P_Glyma.11G141800.Wm82.a2.v1"ret[["N_P_Glyma.19G168700.Wm82.a2.v1"]]# Below are the same three examples, only now using the # SummarizedExperiment (i.e. dataSE) object as input. # Example 1: Create litre plots for each of the 61 genes with FDR < 1e-10. # Examine the first plot (gene "N_P_Glyma.19G168700.Wm82.a2.v1") if (FALSE) { data(se_soybean_ir_sub) assay(se_soybean_ir_sub) <- log(as.data.frame(assay(se_soybean_ir_sub))+1) ret <- plotLitre(dataSE = se_soybean_ir_sub, threshVal = 1e-10, saveFile = FALSE) length(ret) names(ret)[1] ret[[1]] } # Example 2: Create litre plots for each of the five most significant genes # (low FDR values). View plot for gene "N_P_Glyma.19G168700.Wm82.a2.v1". if (FALSE) { geneList <- as.data.frame(rowData(se_soybean_ir_sub)) %>% arrange(N_P.FDR) %>% filter(row_number() <= 5) geneList <- geneList[,1] ret <- plotLitre(dataSE = se_soybean_ir_sub, geneList = geneList, pointColor = "deeppink") names(ret) ret[["N_P_Glyma.19G168700.Wm82.a2.v1"]] } # Example 3: Create one litre plot for each of the five most significant # genes (low FDR values). View the plot for gene # "N_P_Glyma.19G168700.Wm82.a2.v1". Use points instead of the default # hexagons as the background. if (FALSE) { ret <- plotLitre(dataSE = se_soybean_ir_sub, geneList = geneList, pointColor = "deeppink", option = "allPoints") names(ret) ret[["N_P_Glyma.19G168700.Wm82.a2.v1"]] }